| dyads_test_vs_ctrl_m1_shift3 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=12; shift=3; ncol=21; ---myaAAAACCCwa------
; Alignment reference
a 0 0 0 140 94 247 366 422 420 395 4 8 9 149 207 0 0 0 0 0 0
c 0 0 0 134 141 91 12 11 9 29 437 424 428 88 92 0 0 0 0 0 0
g 0 0 0 64 83 38 31 5 4 3 2 11 4 59 57 0 0 0 0 0 0
t 0 0 0 111 131 73 40 11 16 22 6 6 8 153 93 0 0 0 0 0 0
|
| AT1G76870_MA1367.1_JASPAR_shift2 (AT1G76870:MA1367.1:JASPAR) |
 |
0.769 |
0.659 |
6.371 |
0.917 |
0.916 |
4 |
1 |
4 |
2 |
2 |
2.600 |
1 |
; dyads_test_vs_ctrl_m1 versus AT1G76870_MA1367.1_JASPAR (AT1G76870:MA1367.1:JASPAR); m=1/10; ncol2=14; w=12; offset=-1; strand=D; shift=2; score= 2.6; --awmywAAACCrsww-----
; cor=0.769; Ncor=0.659; logoDP=6.371; NsEucl=0.917; NSW=0.916; rcor=4; rNcor=1; rlogoDP=4; rNsEucl=2; rNSW=2; rank_mean=2.600; match_rank=1
a 0 0 209 218 173 87 287 436 418 448 6 0 175 41 177 229 0 0 0 0 0
c 0 0 94 64 127 173 21 5 0 6 450 452 4 150 71 46 0 0 0 0 0
g 0 0 53 59 65 73 3 5 2 0 0 0 257 222 53 35 0 0 0 0 0
t 0 0 100 115 91 123 145 10 36 2 0 4 20 43 155 146 0 0 0 0 0
|
| AT1G72740_MA1353.1_JASPAR_shift4 (AT1G72740:MA1353.1:JASPAR) |
 |
0.817 |
0.562 |
7.184 |
0.911 |
0.913 |
2 |
2 |
3 |
3 |
3 |
2.600 |
2 |
; dyads_test_vs_ctrl_m1 versus AT1G72740_MA1353.1_JASPAR (AT1G72740:MA1353.1:JASPAR); m=2/10; ncol2=15; w=11; offset=1; strand=D; shift=4; score= 2.6; ----cctAAACCCTAAwyc--
; cor=0.817; Ncor=0.562; logoDP=7.184; NsEucl=0.911; NSW=0.913; rcor=2; rNcor=2; rlogoDP=3; rNsEucl=3; rNSW=3; rank_mean=2.600; match_rank=2
a 0 0 0 0 62 69 59 214 282 282 10 0 0 0 289 242 138 9 37 0 0
c 0 0 0 0 154 159 46 3 3 0 281 291 291 0 0 0 41 129 157 0 0
g 0 0 0 0 23 34 51 22 2 9 0 0 0 0 1 49 2 16 27 0 0
t 0 0 0 0 52 29 135 52 4 0 0 0 0 291 1 0 110 137 70 0 0
|
| TBP3.ampDAP_M0621_AthalianaCistrome_rc_shift0 (TBP3.ampDAP:M0621:AthalianaCistrome_rc) |
 |
0.793 |
0.501 |
0.448 |
0.920 |
0.924 |
3 |
4 |
10 |
1 |
1 |
3.800 |
3 |
; dyads_test_vs_ctrl_m1 versus TBP3.ampDAP_M0621_AthalianaCistrome_rc (TBP3.ampDAP:M0621:AthalianaCistrome_rc); m=3/10; ncol2=19; w=12; offset=-3; strand=R; shift=0; score= 3.8; maamhhtaARCCCTAAwyy--
; cor=0.793; Ncor=0.501; logoDP=0.448; NsEucl=0.920; NSW=0.924; rcor=3; rNcor=4; rlogoDP=10; rNsEucl=1; rNSW=1; rank_mean=3.800; match_rank=3
a 126 140 138 102 76 73 50 167 197 186 11 5 0 0 251 231 93 23 51 0 0
c 64 39 30 66 96 83 37 4 2 3 239 237 239 18 3 1 57 71 95 0 0
g 21 48 33 26 19 23 38 35 2 66 1 3 13 5 1 21 9 7 24 0 0
t 44 28 54 61 64 76 130 49 54 0 4 10 3 232 0 2 96 154 85 0 0
|
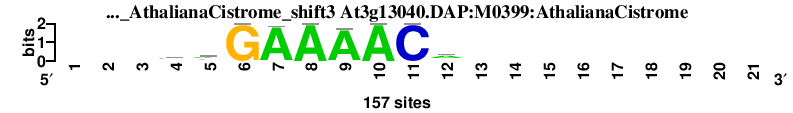
| At3g13040.DAP_M0399_AthalianaCistrome_shift3 (At3g13040.DAP:M0399:AthalianaCistrome) |
 |
0.715 |
0.536 |
6.199 |
0.877 |
0.865 |
9 |
3 |
5 |
10 |
9 |
7.200 |
9 |
; dyads_test_vs_ctrl_m1 versus At3g13040.DAP_M0399_AthalianaCistrome (At3g13040.DAP:M0399:AthalianaCistrome); m=9/10; ncol2=9; w=9; offset=0; strand=D; shift=3; score= 7.2; ---awGAAAACa---------
; cor=0.715; Ncor=0.536; logoDP=6.199; NsEucl=0.877; NSW=0.865; rcor=9; rNcor=3; rlogoDP=5; rNsEucl=10; rNSW=9; rank_mean=7.200; match_rank=9
a 0 0 0 76 72 0 154 157 150 157 0 88 0 0 0 0 0 0 0 0 0
c 0 0 0 20 10 0 0 0 0 0 157 23 0 0 0 0 0 0 0 0 0
g 0 0 0 24 25 157 3 0 7 0 0 13 0 0 0 0 0 0 0 0 0
t 0 0 0 37 50 0 0 0 0 0 0 33 0 0 0 0 0 0 0 0 0
|




